API Basics
In this tutorial, we will cover basic functionalities supported by lineapy using simple examples.
Table of Contents
You can ignore # NBVAL_* comments in certain cell blocks. They are for passing unit tests only, which we do to make sure the examples are always functional as we update the codebase.
[1]:
# NBVAL_IGNORE_OUTPUT
import os
import lineapy
import pandas as pd
import matplotlib.pyplot as plt
No .lineapy folder found. Creating a new folder in /Users/sangyoonpark directory.
Exploring data
Let’s load the toy data to use.
[2]:
# NBVAL_IGNORE_OUTPUT
# Load data
df = pd.read_csv("https://raw.githubusercontent.com/LineaLabs/lineapy/main/examples/tutorials/data/iris.csv")
[3]:
# View data
df
[3]:
| sepal.length | sepal.width | petal.length | petal.width | variety | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Setosa |
| ... | ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | Virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | Virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | Virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | Virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | Virginica |
150 rows × 5 columns
Now, we might be interested in seeing if the data reflects differences between iris types. Let’s compare their petal traits.
[4]:
# Plot petal length/width by iris type
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
df.boxplot("petal.length", "variety", ax=ax[0])
df.boxplot("petal.width", "variety", ax=ax[1])
plt.show()
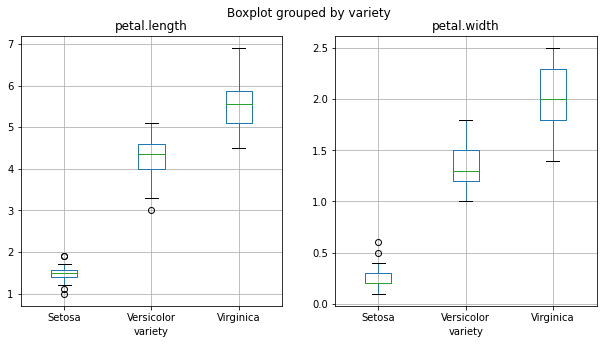
Overall, we observe noticeable differences between iris types, especially between Setosa and Virginica. Let’s quantify differences between the two types.
[5]:
# Calculate averages for Setosa
avg_length_setosa = df.query("variety == 'Setosa'")["petal.length"].mean()
avg_width_setosa = df.query("variety == 'Setosa'")["petal.width"].mean()
# Calculate averages for Virginica
avg_length_virginica = df.query("variety == 'Virginica'")["petal.length"].mean()
avg_width_virginica = df.query("variety == 'Virginica'")["petal.width"].mean()
# Calculate differences
diff_avg_length = avg_length_setosa - avg_length_virginica
diff_avg_width = avg_width_setosa - avg_width_virginica
[6]:
# NBVAL_IGNORE_OUTPUT
# View result
print("Difference in average length:", diff_avg_length)
print("Difference in average width:", diff_avg_width)
Difference in average length: -4.09
Difference in average width: -1.7800000000000002
Storing an artifact with save()
Say we are particularly interested in tracking the average length difference between Setosa and Virginica. For instance, we might want to use this variable later for population-level modeling of the two species.
The save() method allows us to store a variable’s value and history as a data type called LineaArtifact. Note that LineaArtifact holds more than the final state of the variable — it also captures the complete development process behind the variable, which allows for full reproducibility. For more information about artifacts in LineaPy, please check the Concepts section.
The method requires two arguments: the variable to save and the string name to save it as. It returns the saved artifact.
[7]:
# Store the variable as an artifact
length_artifact = lineapy.save(diff_avg_length, "iris_diff_avg_length")
# Check object type
print(type(length_artifact))
<class 'lineapy.graph_reader.apis.LineaArtifact'>
LineaArtifact object has two key APIs:
.get_value()returns value of the artifact, e.g., an integer or a dataframe.get_code()returns minimal essential code to create the value
Hence, for the current artifacts, we see:
[8]:
# Check the value of the artifact
print(length_artifact.get_value())
-4.09
☝️ ☝️ ☝️ [FEATURE] Retrieve artifact value ☝️ ☝️ ☝️
[9]:
# Check minimal essential code to generate the artifact
print(length_artifact.get_code())
import pandas as pd
df = pd.read_csv(
"https://raw.githubusercontent.com/LineaLabs/lineapy/main/examples/tutorials/data/iris.csv"
)
avg_length_setosa = df.query("variety == 'Setosa'")["petal.length"].mean()
avg_length_virginica = df.query("variety == 'Virginica'")["petal.length"].mean()
diff_avg_length = avg_length_setosa - avg_length_virginica
☝️ ☝️ ☝️ [FEATURE] Retrieve cleaned artifact code ☝️ ☝️ ☝️
Note that irrelevant code has been stripped out (e.g., operations relating to diff_avg_width only).
Note: If you want, you can retrieve the artifact’s full original code with artifact.get_session_code().
Listing artifacts with catalog()
Of course, with time passing, we may not remember what artifacts we saved and under what names. The catalog() method allows us to see the list of all previously saved artifacts, like so:
[10]:
# NBVAL_IGNORE_OUTPUT
# List all saved artifacts
lineapy.catalog()
[10]:
iris_diff_avg_length:0 created on 2022-05-17 15:53:09.538389
☝️ ☝️ ☝️ [FEATURE] Retrieve all artifacts ☝️ ☝️ ☝️
Note that the catalog records each artifact’s creation time, which means that multiple versions can be stored under the same artifact name. Hence, if we save iris_diff_avg_length artifact again, we get:
[11]:
# NBVAL_IGNORE_OUTPUT
# Save the same artifact again
lineapy.save(diff_avg_length, "iris_diff_avg_length")
# List all saved artifacts
lineapy.catalog()
[11]:
iris_diff_avg_length:0 created on 2022-05-17 15:53:09.538389
iris_diff_avg_length:1 created on 2022-05-17 15:53:11.370624
Retrieving an artifact with get()
We can retrieve any stored artifact using the get() method. This comes in handy when we work across multiple sessions/phases of a project (or even across different projects) as we can easily build on the previous work.
For example, say we have done other exploratory analyses and are finally starting our work on population-level modeling. This is likely done in a new Jupyter notebook (possibly in a different subdirectory) and we need an easy way to load artifacts from our past work. We can use the get() method for this.
The method takes the string name of the artifact as its argument and returns the corresponding artifact, like so:
[12]:
# Retrieve a saved artifact
length_artifact2 = lineapy.get("iris_diff_avg_length")
# Confirm the artifact holds the same value and code as before
print(length_artifact2.get_value())
print(length_artifact2.get_code())
-4.09
import pandas as pd
df = pd.read_csv(
"https://raw.githubusercontent.com/LineaLabs/lineapy/main/examples/tutorials/data/iris.csv"
)
avg_length_setosa = df.query("variety == 'Setosa'")["petal.length"].mean()
avg_length_virginica = df.query("variety == 'Virginica'")["petal.length"].mean()
diff_avg_length = avg_length_setosa - avg_length_virginica
By default, the get() method retrieves the latest version of the given artifact. To retrieve a particular version of the artifact, we can specify the value of the optional argument version, like so:
[13]:
# NBVAL_IGNORE_OUTPUT
# Get version info of the retrieved artifact
desired_version = length_artifact2.version
# Check the version info
print(desired_version)
print(type(desired_version))
1
<class 'int'>
[14]:
# NBVAL_IGNORE_OUTPUT
# Retrieve the same version of the artifact
length_artifact3 = lineapy.get("iris_diff_avg_length", version=desired_version)
# Confirm the right version has been retrieved
print(length_artifact3.name)
print(length_artifact3.version)
iris_diff_avg_length
1
Using artifacts to build pipelines
Say we are now also interested in using the average width difference in our population-level modeling, in which case we will store it as an artifact too:
[15]:
# Store the width variable as an artifact too
width_artifact = lineapy.save(diff_avg_width, "iris_diff_avg_width")
Now consider the case where our source data (i.e. iris.csv) gets updated. Moreover, the update is not a one-time event; the data is planned to be updated on a regular basis as new samples arrive.
Since the iris_diff_avg_length and iris_diff_avg_width artifact were derived from the iris.csv data, this means that we need to rerun each artifact’s code lest its value be stale. Given the recurring updates in the source data, we may want to build and schedule a pipeline to automatically rerun the code of both artifacts on a regular basis.
Having the complete development process captured in each artifact, LineaPy makes it easy for us to to turn these two artifacts into a deployable pipeline. For instance, Airflow is a popular tool for pipeline building and management, and we can turn the artifacts into a set of files that can be deployed as an Airflow DAG, like so:
[16]:
# NBVAL_IGNORE_OUTPUT
# Build an Airflow pipeline with both length and width artifacts
lineapy.to_pipeline(
artifacts=[length_artifact.name, width_artifact.name],
pipeline_name="demo_pipeline",
framework="AIRFLOW",
output_dir="output/00_api_basics/demo_pipeline/",
)
Pipeline source generated in the directory: output/00_api_basics/demo_pipeline
Generated python module demo_pipeline.py
Generated Dockerfile demo_pipeline_Dockerfile
Generated requirements file demo_pipeline_requirements.txt
Added Airflow DAG named demo_pipeline_dag. Start a run from the Airflow UI or CLI.
[16]:
PosixPath('output/00_api_basics/demo_pipeline')
where
artifactsis the list of artifact names to be used for the pipelinepipeline_nameis the name of the pipelineoutput_diris the location to put the files for running the pipelineframeworkis the name of orchestration framework to use (currently supportsSCRIPTSandAIRFLOW)
And we see the following files have been generated:
[17]:
# NBVAL_IGNORE_OUTPUT
# Check the generated files for running the pipeline
os.listdir("output/00_api_basics/demo_pipeline/")
[17]:
['demo_pipeline_requirements.txt',
'demo_pipeline_Dockerfile',
'demo_pipeline_dag.py',
'demo_pipeline.py']
where
[PIPELINE-NAME].pycontains the artifact’s cleaned-up code packaged as a function[PIPELINE-NAME]_dag.pyuses the packaged function to define the pipeline[PIPELINE-NAME]_requirements.txtlists dependencies for running the pipeline[PIPELINE-NAME]_Dockerfilecontains commands to set up the environment to run the pipeline
Specifically, we have demo_pipeline.py looking as follows:
[18]:
# NBVAL_IGNORE_OUTPUT
%cat output/00_api_basics/demo_pipeline/demo_pipeline.py
import pickle
def iris_diff_avg_length():
import pandas as pd
df = pd.read_csv(
"https://raw.githubusercontent.com/LineaLabs/lineapy/main/examples/tutorials/data/iris.csv"
)
avg_length_setosa = df.query("variety == 'Setosa'")["petal.length"].mean()
avg_length_virginica = df.query("variety == 'Virginica'")["petal.length"].mean()
diff_avg_length = avg_length_setosa - avg_length_virginica
pickle.dump(
diff_avg_length,
open("/Users/sangyoonpark/.lineapy/linea_pickles/CazbEiW", "wb"),
)
def iris_diff_avg_width():
import pandas as pd
df = pd.read_csv(
"https://raw.githubusercontent.com/LineaLabs/lineapy/main/examples/tutorials/data/iris.csv"
)
avg_width_setosa = df.query("variety == 'Setosa'")["petal.width"].mean()
avg_width_virginica = df.query("variety == 'Virginica'")["petal.width"].mean()
diff_avg_width = avg_width_setosa - avg_width_virginica
width_artifact = pickle.dump(
diff_avg_width, open("/Users/sangyoonpark/.lineapy/linea_pickles/l5PX1bp", "wb")
)
We can see that LineaPy used artifacts to automatically 1) clean up their code to retain only essential operations and 2) package the cleaned-up code into importable functions.
And we see demo_pipeline_dag.py automatically composing an Airflow DAG with these functions:
[19]:
# NBVAL_IGNORE_OUTPUT
%cat output/00_api_basics/demo_pipeline/demo_pipeline_dag.py
import os
import demo_pipeline
from airflow import DAG
from airflow.operators.python_operator import PythonOperator
from airflow.utils.dates import days_ago
default_dag_args = {"owner": "airflow", "retries": 2, "start_date": days_ago(1)}
dag = DAG(
dag_id="demo_pipeline_dag",
schedule_interval="*/15 * * * *",
max_active_runs=1,
catchup=False,
default_args=default_dag_args,
)
iris_diff_avg_length = PythonOperator(
dag=dag,
task_id="iris_diff_avg_length_task",
python_callable=demo_pipeline.iris_diff_avg_length,
)
iris_diff_avg_width = PythonOperator(
dag=dag,
task_id="iris_diff_avg_width_task",
python_callable=demo_pipeline.iris_diff_avg_width,
)
These files, once placed in the location that Airflow expects (usually dag/ under Airflow’s home directory), should let us immediately execute the pipeline from the UI or CLI.
For a more detailed illustration of pipeline building, please check this tutorial.
Recap
In this tutorial, we learned basic functionalities of LineaPy including how to save, browse, and retrieve an artifact. We saw that a LineaPy artifact stores not only the value of a variable but also its full development code. This then helps to automate time-consuming, manual steps in a data science workflow such as code cleanup and pipeline building, hence helping data scientists move faster towards productionization and impact.